Introduction to modality XPLR#
Methylation data has diverse applications from basic science to translational research and test development. The modality XPLR analysis software is a fast and scalable command line interface (CLI) toolkit, for the analysis of methylation data that is derived from 5-base (duet multiomics solution +modC) and 6-base (duet multiomics solution evoC) genomes. The software leverages multi-core, out-of-memory processing to enable efficient computation on a standard laptop or HPC; allowing users to work on large datasets with ease.
The modality XPLR analysis software includes tools for exploratory analyses (e.g. computation and plotting of methylation statistics and differentially methylated regions (DMRs)), as well as supporting downstream analysis (e.g. feature extraction and export functions). Designed for efficiency and ease of use, it enables you to rapidly transition from raw data to actionable insights and publication-ready visualisations.
The core data file used by modality XPLR is the Zarr store, which is an array format that includes methylation and coordinate data, produced by the (duet Software).
The software also allows export to community-supported file types for analysis with other informatics tools.
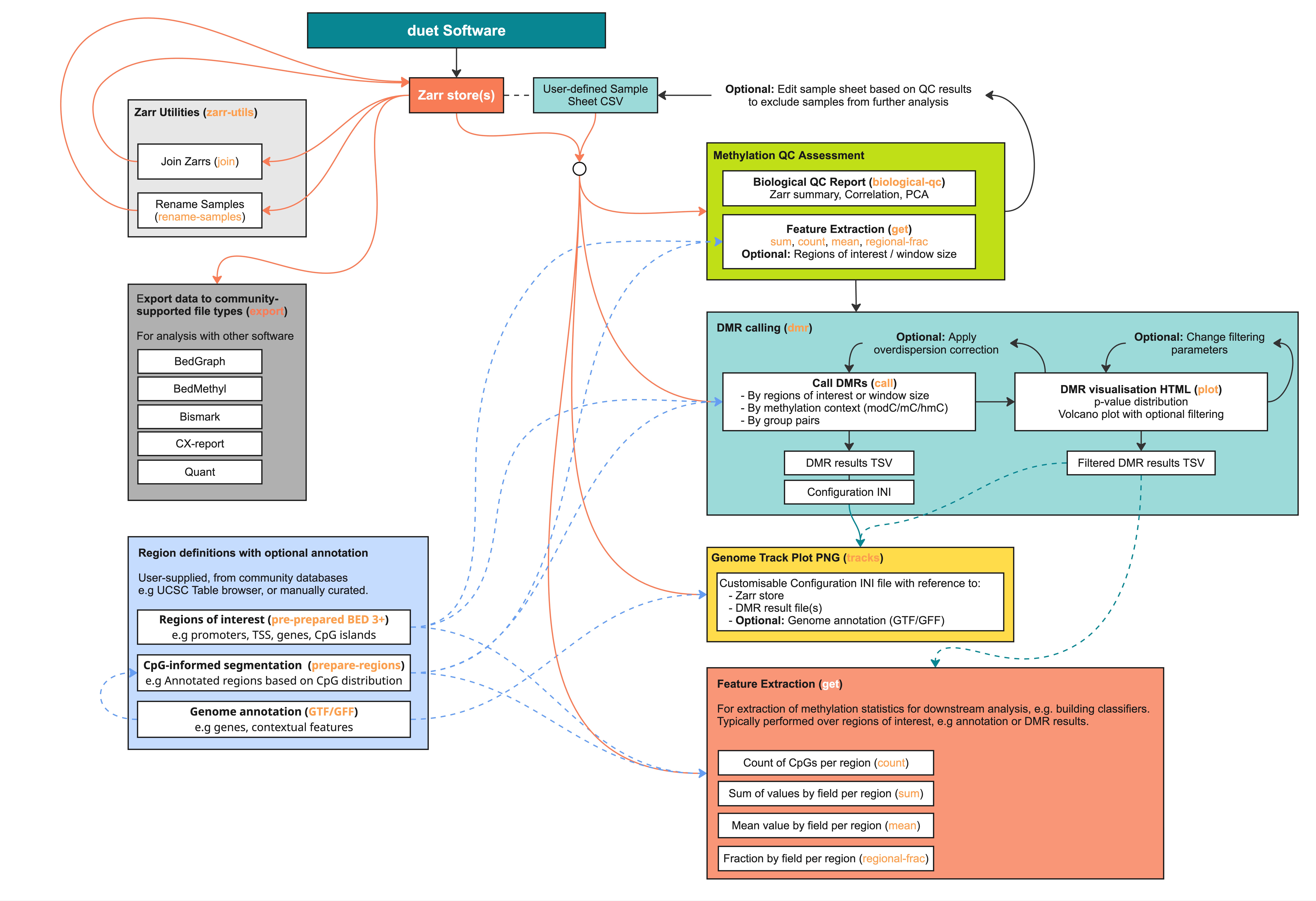
Figure 1 Schematic of the modality XPLR analysis software and example workflows. The Zarr store is generated by the duet Software, and can be used as input to modality XPLR tools.#
User documentation contents#
- Getting started
- Installation of modality XPLR
- Authenticate modality XPLR
- Core Workflow
- Interpreting Core Workflow outputs
- Advanced analysis workflow
- Toolkit
- Preparing for analysis
- Handling Zarr stores
- Prepare annotated regions
- Biological QC
- Calling differentially methylated regions (DMRs)
- Plotting differentially methylated regions (DMRs)
- Generate genomic track plots
- Extracting methylation statistics
- Audit trail
- Example workflow
- Overview of the workflow
- 1. Assess methylation data quality
- 2. Checking CpG context depth across the regions of interest
- 3. Call differentially methylated regions (DMRs) in late stage disease
- 4. Filtering DMRs to identify statistically significant changes
- 5. Use biological priors to call DMRs in early stage disease
- 6. Visualisation of key early-stage 5hmC DMRs
- 7. Place methylation changes (DMRs) into genomic context
- 8. Generate feature sets for building classifiers
- Conclusion
- FAQs and troubleshooting
How to use this guide#
Support links#
For any questions or issues, please contact your biomodal support team at support@biomodal.com.
Release notes#
A summary of recent changes to the software is provided below. For a full list of changes, please refer to the published release notes PDF by clicking on the version number.
Release version |
Date |
Description of changes |
|---|---|---|
2025-Nov-17 |
|
|
2025-Oct-03 |
|
|
2025-Sep-22 |
|
|
2025-Aug-04 |
|
|
2025-Jun-30 |
Initial beta release. |
