biomodal Data Interpretation: Quick Start Guide#
This quick start guide gives a concise overview of how to use our Data Interpretation Guide. It outlines the key steps to assess your data quality and begin exploring your duet pipeline outputs, depending on the level of analysis you want to achieve, whether you are a lab scientist or a bioinformatician. The full Data Interpretation Guide is comprehensive, you don’t need to read it all at once. Instead, use the sections that are most relevant to your role and the depth of analysis you’re aiming for.
Who Is This For?#
This guide is for anyone who has run the duet pipeline and wants to understand how to interpret the output data. It is particularly useful for:
🔬 Lab Scientist: You want to understand what the data says about your biological samples.
🧑💻 Bioinformatician: You want to explore, process, or integrate the data into downstream analyses.
Overview: From Data to Insight#
Note
The diagram below shows the main files the duet pipeline produces, and how they can be used to derive biological insights. The duet pipeline produces additional optional output files that are not listed in the diagram.
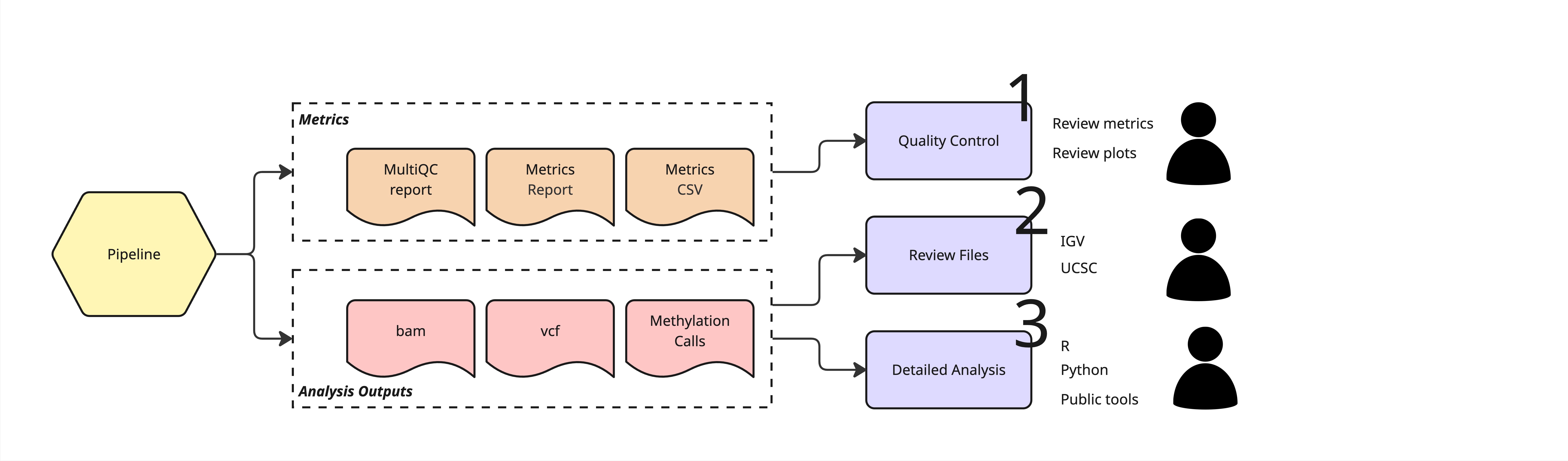
Review the metrics and plots generated by duet to make sure your data is of high quality.
Explore individual output files with standard tools to get a feel for your data.
Use the output files to derive biological insights, such as differential methylation or integration with other omics data.
Step-by-Step#
For Lab Scientists#
Check Your Assay Type - duet +modC or duet evoC? See assay overview
Review technical QC measures - Understand how duet assesses accuracy. Summary metrics
Open the Summary Report - Review key metrics. Metrics summary and MultiQC report
Explore Methylation Calls - Find the methylation output files that suit your needs. Epigenetic Quantification
For Bioinformaticians#
Locate Output Files - BAM, BED, TSV, JSON. File formats
Dive into the Metrics - Use summary files or raw data. Metrics analysis
Visualise your alignment files - Using IGV, R, Python. Alignment output files
Explore Epigenetic Quantification - Using one of the output files. Quantification
